
Congratulations to the CBU director Nathalie Reuter and group leader Sushma Grellscheid on getting the grant for their project funded by the Research Council of Norway.
For more information, please check here.

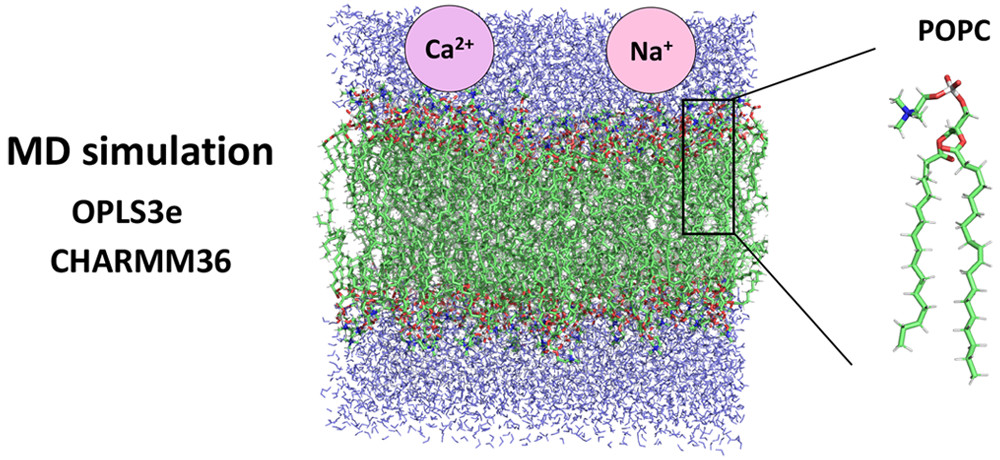
Our group leader Markus Miettinen has his paper out with the title: Structure of POPC Lipid Bilayers in OPLS3e Force Field. Read here.

Congratulations to the CBU director Nathalie Reuter and group leader Sushma Grellscheid on getting the grant for their project funded by the Research Council of Norway.
For more information, please check here.

On August 24th Muhammad Ammar Malik successfully defended his PhD thesis with the title: Machine learning approaches for high-dimensional genome-wide association studies
Genome-wide association studies (GWAS) aim to find statistical associations between genetic variants and traits of interests. The genetic variants that explain a lot of variation in genome-wide gene expression may lead to confounding in expression quantitative trait loci (eQTL) analyses. To account for these confounding factors, we proposed LVREML, a method conceptually analogous to estimating fixed and random effects in linear mixed models (LMM). We showed that the maximum-likelihood latent variables can always be chosen orthogonal to the known factors (such genetic variants). This indicates that the maximum likelihood variables explain the sample covariances that is not already explained by the genetic variants in the model.
For identifying which traits are affected by the identified genetic variants, we need to reverse the functional relation between genotypes and traits. In this regard, multi-trait approaches are more advantageous than studying the traits individually. The multi-trait approaches benefit from increased power from considering cross-trait covariances and reduced multiple testing burden because a single test is needed to test for associations to a set of traits. Therefore, we analyzed various machine learning methods (ridge regression, Naive Bayes/independent univariate correlation, random forests and support vector machines) for reverse regression in multi-traitGWAS, using genotypes, gene expression data and ground-truth transcriptional regulatory networks from the DREAM5 SysGen Challenge and from a cross between two yeast strains to evaluate methods.
We then extended the above approach to human dataset. We used the genotype and brain-imaging features extracted from the MRIs obtained from the ADNI database. Our results showed that the genotype prediction performance varied across genetic variants. This helped in identifying genomic regions that are associated with high number of traits in high-dimensional phenotypic data. We also observed that the feature coefficients of fitted machine learning models correlated with the strength of association between variants and traits. Our results also showed that non-linear machine learning methods like random forests identified genetic variants distinct from the linear methods. In particular, we observed that random forest was able to identify single-nueclotide-polymorphisms (SNPs) that were distinct from the ones identified by ridge and lasso regression. Further analysis showed that the identified SNPs belonged to genes previously associated with brain-related disorders.
Publications
Restricted maximum-likelihood method for learning latent variance components in gene expression data with known and unknown confounders
(https://doi.org/10.1093/g3journal/jkab410)
Malik, M. A., & Michoel, T.
G3, 12(2), jkab410.
High-dimensional multi-trait GWAS by reverse prediction of genotypes
(https://doi.org/10.48550/arXiv.2111.00108)
Malik, M. A., Ludl, A. A., & Michoel, T.
In 2021 International Meeting on Computational Intelligence Methods for Bioinformatics and Biostatistics. Springer, Cham.
rfPhen2Gen: A machine learning based association study of brain imaging phenotypes to genotypes
(https://doi.org/10.48550/arXiv.2204.00067)
Malik, M. A., Lundervold, A. S., & Michoel, T.
Under review at Bioinformatics Advances.

Bram Danneels is our new Postdoctoral Fellow who has joined the CBU as part of the EBP-Nor project to help develop tools and pipelines for the efficient and high-throughput assembly and annotation of the eukaryotic target species, under supervision of Michael Dondrup and professor Inge Jonassen. Bram got his PhD in Bioinformatics at Ghent University. After his PhD, Bram did a short Post-Doc with prof. Aurélien Carlier at the LIPME where he analysed RNA-sequencing data from the Dioscorea-Orrella symbiosis and investigated the Dioscorea sansibarensis plant genome.
During the PhD Bram worked in the lab of prof. Aurélien Carlier (part of the Laboratory of Microbiology (LMG) at Ghent University), where he studied leaf symbiosis from an evolutionary perspective. They investigated mainly the symbiosis between the Zanzibar yam (Dioscorea sansibarensis) and its bacterial leaf symbiont Orrella dioscoreae. Bram studied the diversity and evolution of the symbiont’s genome by using comparative genomics on metagenome-assembled genomes from plants collected from different geographical regions. He also studied leaf symbiosis between the plant families Rubiaceae and Primulaceae with bacteria from the Burkholderiaceae family. During his PhD, Bram spent 2 years at the Laboratory for Interactions between Plants, Microbes, and the Environment (LIPME) in Toulouse, France.